The SARS-CoV-2 variant B.1.1.7 virus, first detected in the UK in late October 2020, has become the dominant strain in the UK, throughout most of Europe and India, and is now the most frequent virus isolated in the United States. The B.1.1.7 variant is more infectious than the SARS-CoV-2 strains that circulated earlier in the pandemic, is almost twice as deadly, and infects younger people and even children at a higher rate. Many speculate that the current surge of infections throughout Europe, the United States, and possibly India can be attributed, at least in part, to the increased efficiency of transmission of the B.1.1.7 virus.
The hope is that the current generation of Covid-19 vaccines, all of which are based on the spike protein of the original Wuhan strain, will be effective against the B.1.1.7 strain. This hope is based on the observation that antibodies from plasma of those who receive the vaccine recognize and neutralize the B.1.1.7 virus only slightly less well than they do the original Wuhan strain.
Now comes a troubling report of the discovery of a new variant from Oregon. As of yet, there are only two confirmed isolates in Oregon with this sequence, but the number of undetected cases is likely larger. The variant does not yet have an official designation. I will call it B.1.1.7-O as the variant carries all of the mutations of B.1.1.7 plus an additional six mutations of its own, one in the spike protein, three on the replication complex (ORF1ab), one in the structural protein N, and one in the accessory protein ORF8. This variant arises on the west coast as the B.1.427/9 variants have surged through California, making up over 50% of the cases sequenced.
Of these, the single change in the spike protein, a substitution of the negatively charged amino acid glutamic acid (E) for the positively charged lysine at position 484 (E484K), nicknamed the “EEK!” variant, is of special concern particularly when coupled with the N501Y mutation (asparagine (N) to tyrosine (Y) at amino acid 501). The combination of these two mutations in the receptor-binding domain of the spike protein is the principal cause of vaccine resistance of the Brazilian isolate B.1.1.28.1and the South African variant B.1.351. Together these two mutations are associated with an increased transmission and vaccine resistance. For example, the Astra-Zeneca adenovirus-based vaccine is only 10% effective against the B.1.351 South African variant. The potency of the Moderna and Pfizer vaccines is reduced almost tenfold when tested against the B.1.351 and B.1.1.28.1 variants. The B.1.1.7-O variant is likely to be similarly resistant to the current vaccines. The figure below depicts the full range of B.1.1.7-O’s spike mutations.
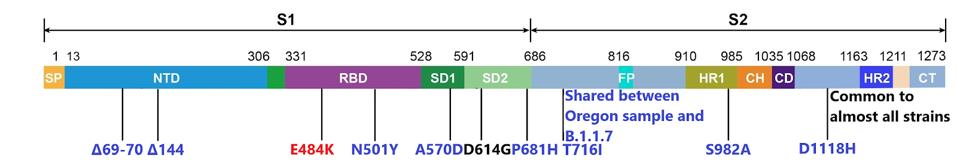
The single additional mutation in the accessory gene ORF8 may also contribute to increased infectivity and/or immune evasion. The mutation at position 68 of ORF8 introduces a stop codon preventing the expression of the protein altogether. This is in addition to a stop codon at position 27 and two amino acid changes at positions 52 and 71 of B.1.1.7. ORF8 is highly antigenic and the stop codon introduces two non-exclusive possibilities. First, ORF8 is eliminated as a key immunogenic epitope, allowing increased immune evasion in the variant. Second, its elimination may remove a factor that attenuates virulence.
Premature termination in ORF8 in SARS-CoV-2 is relatively rare, but other mutations to the accessory protein occur in the B.1.1.28.1, Californian B.1.427/9, New York B.1.526, and Philippine B.1.1.28.3 variants. The selection pressure against the expression of ORF8 seems to be exceptionally powerful. The ORF8 protein is expressed upon infection with the original Wuhan strain and the D614G variantB.1as evidenced by antibodies to the ORF8 with convalescent sera. Is it possible that ORF8 attenuates the virulence of SARS-CoV-2 and that the premature termination increases symptoms in infected hosts?
The four additional mutations that distinguish B.1.1.7-O from B.1.1.7 include one change of a single amino acid in each of the nonstructural proteins, NSP2, the polymerase NSP12, and exonuclease NSP14, and in the structural protein, N. The NSP2 and N mutations are rare, showing up in only a few hundred isolates of B.1.1.7 of the over 300,000 in the GISAID database. The NSP12 and NSP14 mutations, however, are a bit more common, but not lineage-defining. The NSP12 mutation appears in around 28,000 isolates and the NSP14 mutation in around 6,000 isolates, with both below 10% of sequenced B.1.1.7 isolates. The figure below depicts the full range of B.1.1.7-O’s genomic mutations outside of the spike protein.

The detection B.1.1.7-O is troubling for several reasons. The variant may spread rapidly in the United States and may be vaccine-resistant. The variant also may be able to re-infect those infected earlier in the pandemic as is the case for both B.1.351 and B.1.1.28.1 that carry the E484K and N501Y mutations.
The appearance of B.1.1.7-O serves as a warning of the continuing evolution of SARS-CoV-2 from older variants, like B.1.1.7, towards increased transmissibility, virulence, and immune evasion in the United States and elsewhere. Effective public health mitigation measures, surveillance, and rapid deployment of second-generation vaccines seem the only way to contain this ever more dangerous virus.

