A group of seven residents in a Belgian nursing home died after infection with a new variant, all of whom were fully vaccinated according to virologists on the scene. Though the vaccine used in these residents was not made public, it was likely either AstraZeneca or an mRNA vaccine such as Pfizer and Moderna, among those most frequently used in the country.
The new variant, B.1.621, has yet to receive a greek letter designation from the World Health Organization. The variant already accounts for around 2% of coronavirus infections in the United States, and as much as 10% in Miami, according to Carlos Migoya, CEO of Jackson Health System. The variant can be traced back to identification in samples from Colombia in March 2021 and according to the GISAID coronavirus database, the variant has been identified in as many as 28 countries, but potentially more.
Despite not having the official label of Variant of Interest or Concern, it should be noted that B.1.621 has certain characteristics that suggest it may be worth watching, as it could become a variant of interest or concern in the near future. Here we analyze those mutations and summarize what we know about their likely effects on transmissibility, virulence, vaccine evasion, and pathogenesis.
Figure 1 shows both the nonsynonymous nucleotide mutations that result in amino acid changes and synonymous “silent” nucleotide mutations in B.1.621 versus the original Wuhan strain. What follows is a brief tour of the potential impact of the mutations beginning at the 5’ end of the genome and extending to the 3’ end. Altogether, there are 17 nucleotide mutations, of which 13 result in amino acid mutations.
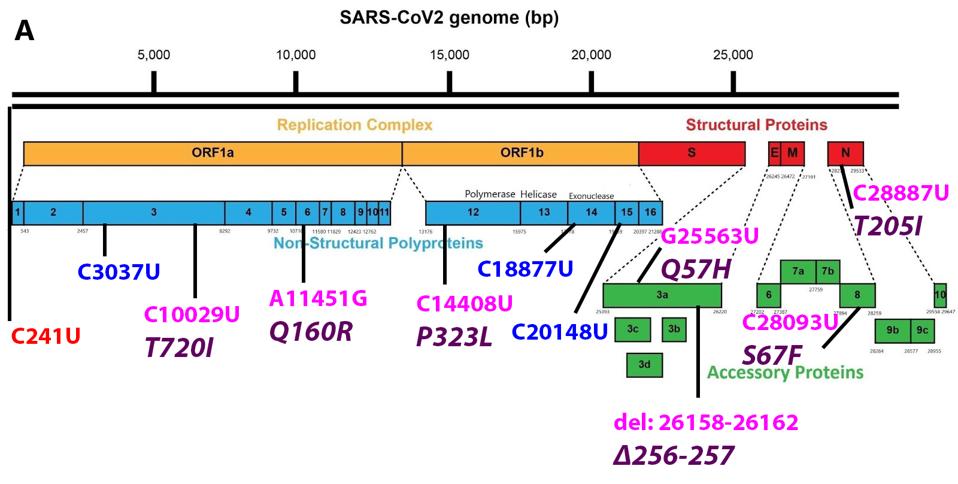

The 5’ most mutation is C241U. This mutation in the untranslated noncoding region, along with P323L in nonstructural protein (NSP) 12 and D614G in the Spike (S) protein, appear in almost all currently circulating strains today. These first appeared in late January and early February 2020 and have since become the dominant strain around the world. Almost all variants of interest or concern harbor these three mutations derived from the early variant. The D614G mutation in the S protein is critical to increased transmissibility and P323L in NSP12 is speculated to increase the replication efficiency of the virus. Here we speculate that the C241U 5’ end mutation may confer increased production of full-length genomic RNA and packaging efficiency. This mutation is located in loop three of the leader sequence of genomic and subgenomic RNAs. We also note this mutation may affect the frequency of translation as it is located 25 nucleotides 5’ to the Orf1ab initiation codon.
The B.1.621 variant has two mutations in NSP3—one silent and one amino acid altering. NSP3 is a multi-domain protein with a number of different functions. C3037U is a synonymous mutation that lies in the Acidic C-terminal (Ac) domain. The next mutation, T720I, lies in the SARS-specific unique domain (SUD). The SUD is involved in the formation of G-quadruplexes, which play a role in the modulation of host cell response to infection. Threonine to isoleucine is a major, polar-to-hydrophobic change, indicating a potentially major contribution to the SUD’s operation.
Q160R is found in NSP6. This mutation is another major change from polar uncharged glutamine to positively charged arginine, which has a potential effect on protein structure and function. NSP6 is involved in a protein involved in the formation of the double-membrane replication vesicle, major inhibition of human immune responses by inhibiting STAT1 and STAT2 phosphorylation, and inhibition TANK binding kinase 1. We speculate that this mutation may contribute to pathogenesis by increased suppression of the immune response allowing the virus additional opportunity to replicate.
C18877U is located in NSP14 and C20148U is located in NSP15. As silent mutations, they may have an effect on the stability of the messenger RNA.
The S protein of B.1.621 carries six common mutations in GISAID sequenced samples.
The most 5’ mutation in the S protein is T95I in the N-terminal domain. The N-terminal domain is a major binding site for monoclonal antibodies and other antiviral treatments. Mutations to this domain have been shown to increase virus escape from these medical interventions. T95I is a change from polar threonine to hydrophobic isoleucine. We speculate this could eliminate a binding site, potentially contributing to immune evasion. The significance of this mutation is heightened by the independent observation of the T95I mutation in the Iota variant of interest.
There are two mutations in the receptor-binding domain. The receptor-binding domain interacts directly with the host receptor ACE2. The mutations in the receptor-binding domain are found in many variants of interest and concern. E484K is reported to increase viral resistance to monoclonal antibodies and convalescent sera. N501Y is consistently noted to highly increase avidity between the receptor-binding domain and ACE2 receptor. Both are present in a number of VOIs and VOCs, including Alpha, Beta, Gamma, Eta, and Iota.
The S protein also carries the P681H mutation. Both this and D614G are associated with enhancing the furin cleavage site in the middle portion of the S protein, which again contributes to highly increased transmissibility. D614G is a major change from negative aspartic acid to uncharged glycine. P681H is uncharged proline to positive arginine, again indicating a significant change. The P681H mutation is notable for its observation in the Alpha variant, the fuel behind the third wave of Covid-19 in late 2020 and early 2021.
The final S protein mutation is D950N. The mutation is located in heptad repeat 1. The change from aspartic acid to asparagine is negative to polar uncharged, indicating a major shift in charge and polarity. Although the exact function of this mutation is unknown, it is also found in the currently pervasive Delta variant, likely indicating some unknown increase in viral infectivity. Below is a comprehensive summary of the B.1.621 S protein and the documented and potential effects of its mutations (Figure 2).

There are two additional mutations in Orf3a. The first mutation is the Q57H. A major change from polar uncharged glutamine to positive histidine, this mutation has also been observed in the Iota variant, as well as B.1.427/429 first identified in California. The second mutation is a deletion from nucleotides 26158 to 26162 is commonly noted, which affects amino acids 256 and 257 and is unique to this variant. Evidence suggests Orf3a is important for viral pathogenesis These mutations may enhance Orf3a efficiency.
There is one mutation in Orf8, S67F. This mutation is unique in relation to VOIs and VOCs, though that does not rule out its potential impact on the protein. Orf8 is a major immune regulator, inhibiting B and T cell recognition and downregulating major histocompatibility complex I (MHC-I). The serine to phenylalanine mutation is polar to hydrophobic, again indicating a major change. We suspect that the S67F mutation could very well improve the immune suppressive nature of Orf8 and the virus as a whole.
Near the 3’ end, the Nucleocapsid (N) protein contains the mutation T205I. Found in the Beta VOC, Eta VOI, and B.1.427/429, this mutation may have a strong impact on the immune suppression conducted by the N protein. In addition to packaging and protecting viral RNA, the N protein has a number of suppressive functions, including antiviral RNAi and INF-β antagonism, cyclin-cyclin dependent kinase inhibition, nonsense-mediated decay inhibition, and late infection immune activation. The N protein is an immune suppressive workhorse and T205I, polar threonine to hydrophobic isoleucine, likely contributes to this efficiency. As this is a significant change in the amino acids, it is worth a significant effort to understand the mutation’s impact on the multifunctional N protein.
Our conclusion from the extensive set of mutations found in this particular variant is that it deserves to be observed closely. We have been informed through private communication that because of the Orf3a deletion, it may not routinely be included in the GISAID catalog. Therefore, the representation of its true frequency in populations around the world is underrepresented. B.1.621 clearly has the potential to be lethal in vaccinated elderly patients and may have the potential to become more transmissible as well. Given the described changes, we can begin to understand why the B.1.621 variant should be considered certainly a variant of interest, if not a variant of concern, and merits much closer observation in the coming weeks and months.

