This is the eighth article in a 15-part series called “Relevance of Immune Suppression by SARS-CoV-2 to Understanding and Controlling the Covid-19 Pandemic,” which will explore an underappreciated but highly significant aspect of SARS-CoV-2 replication. The ability of SARS-CoV-2 to delay, evade, and suppress the immune system has myriad implications for drugs, vaccines, and other aspects of our pandemic response. The first set of pieces in this series are intended for a general audience; the second set, for the medical community; and the third and final set, for biomedical researchers looking for a deeper understanding of variants, how they’re generated, and what we might do to control them. Read parts 1, 2, 3, 4, 5, 6, and 7.
SARS-CoV-2 Multilevel Blockade of Cellular Gene Expression
In this section, I will discuss how SARS-CoV-2 blocks expression of almost all cellular genes and proteins to favor its own messenger RNA and protein synthesis. More specifically I will discuss transcription initiation, messenger RNA splicing and stability, messenger RNA export to the cytoplasm, and protein translation.
Blockage of transcription initiation
The first step in assembling this blockade consists of inhibition of initiation of transcription of a specific messenger RNA species, those which specify type 1 interferons and interferon responsive genes. We will discuss these specific pathways in a later installment of this series.
Splicing inhibition
Next comes the inhibition of splicing. The nonstructural protein NSP16 inhibits the splicing of cellular RNAs. Primary nuclear gene transcripts are in general rapidly degraded, unless spliced and exported. While most cellular RNAs are spliced, SARS-CoV-2 messenger RNAs do not require nuclear splicing. NSP16 allows for preferential production of viral RNAs and proteins over their cellular counterparts.
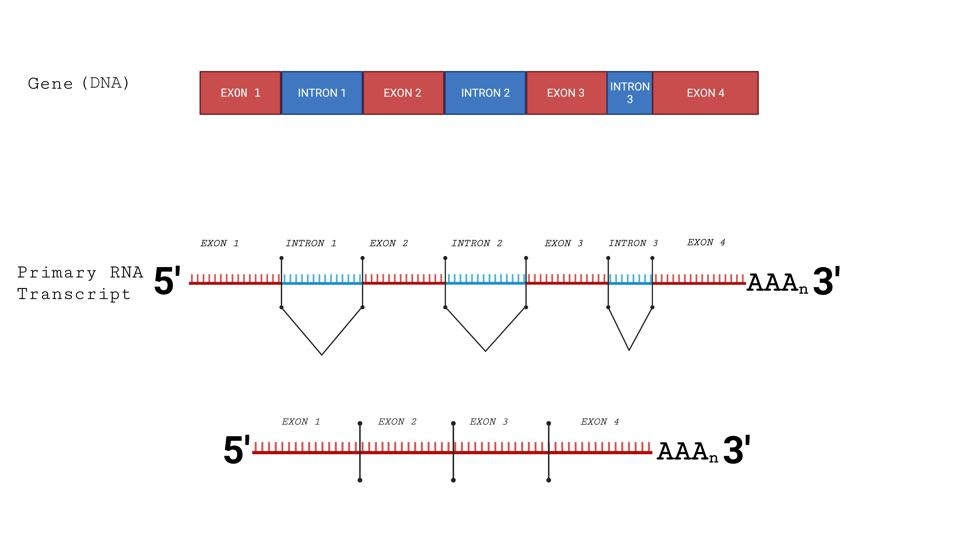
Splicing of cellular messenger RNAs.Figure 1: Splicing of cellular messenger RNAs. This process occurs within the nucleus. Cellular messenger RNAs are made in the nucleus and exported to the cytoplasm. SARS-CoV-2 messenger RNAs are not spliced, made in the cytoplasm, and not found inACCESS HEALTH INTERNATIONAL…Insert Text Above
Following transcription, messenger RNAs must be spliced before they can be exported and translated into proteins. The spliceosome, a protein-RNA complex, catalyzes the splicing reaction by removing introns, a specific type of nucleotide sequence, from cellular messenger RNA transcripts (Figure 1). Central to the action of the spliceosome are two RNA sequences, designated U1 and U2, which bind to nascent RNA to initiate the splicing process.
A recent study found that NSP16 binds to the spliceosome RNA. To be more precise, it binds to both the U1 and U2 sites of the spliceosome RNA, which play a critical role in recognition of the ends of the introns to be removed. Binding of NSP16 prevents the spliceosome from recognizing the corresponding intron sequences and blocks splicing altogether (Figure 2). The net result is nascent cellular messenger RNAs are not matured. In confirmation the authors also reported a marked increase in intron retention in cells infected by SARS-CoV-2, evidence that many transcripts went unspliced due to NSP16-mediated interference.
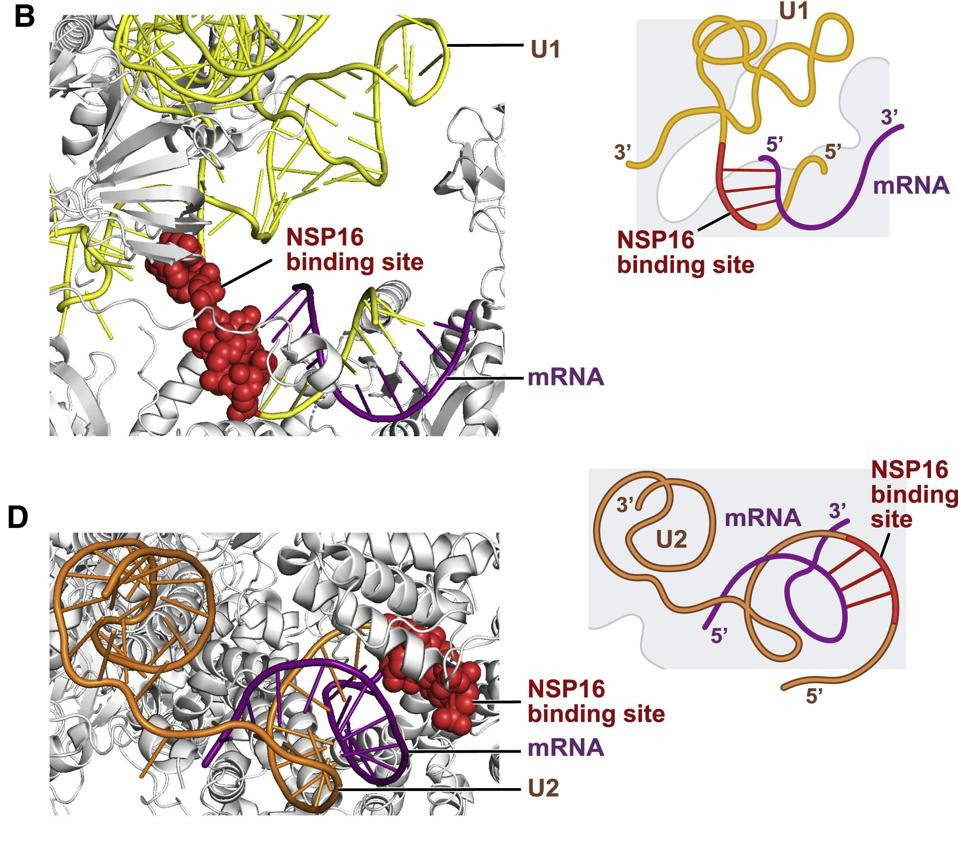
NSP16 Binds to U1 and U2 at Their mRNA Recognition SitesFigure 2: NSP16 Binds to U1 and U2 at Their mRNA Recognition Sites“SARS-COV-2 DISRUPTS SPLICING, TRANSLATION, AND PROTEIN TRAFFICKING TO SUPPRESS HOST DEFENSES”
There are several important consequences of degrading messenger RNA. First and foremost, cellular signals that might be newly initiated to protect the cell from infection are never made. This includes any protein, such as interferons, or other proteins which the cell may use to defend itself, meaning the virus has a free pass to continue replicating. It also means the infected cell cannot make new proteins that will signal to nearby cells or the immune system that the cell is infected.
Replication of viruses may also be inhibited by several different classes of cellular RNA. One of the most intensively investigated are microRNAs. MicroRNAs are small, non-coding RNAs that help cells control gene expression. They act by binding to corresponding sites on viral RNA, leading to its degradation. Blocking splicing and subsequent degradation of nuclear messenger RNA is very likely to inhibit the synthesis and activity of microRNAs as well and any other RNA species of which there are several that are postulated to have antiviral capabilities. Blocking splicing is therefore a critical step in ensuring a favorable environment for viral replication.
If cellular messages that have been initiated and transcribed cannot be spliced, they are degraded, reducing the total number of cellular messenger RNAs. Not only that, but because microRNAs are carved from full length messenger RNAs, I speculate this substantially downregulates the cellular production of microRNAs as well, which might otherwise have antiviral activity.
Blockage of nuclear export
If NSP16-mediated splicing inhibition is not complete or messenger RNA contains no introns, there is still a possibility they will be exported. However another block occurs at the nuclear pore (Figure 3). For messenger RNA to be translated it must be shuttled out of the nucleus and into the cytoplasm via a nuclear pore complex, an elaborate structure that mediates both exit and entry of macromolecules like proteins and RNA.
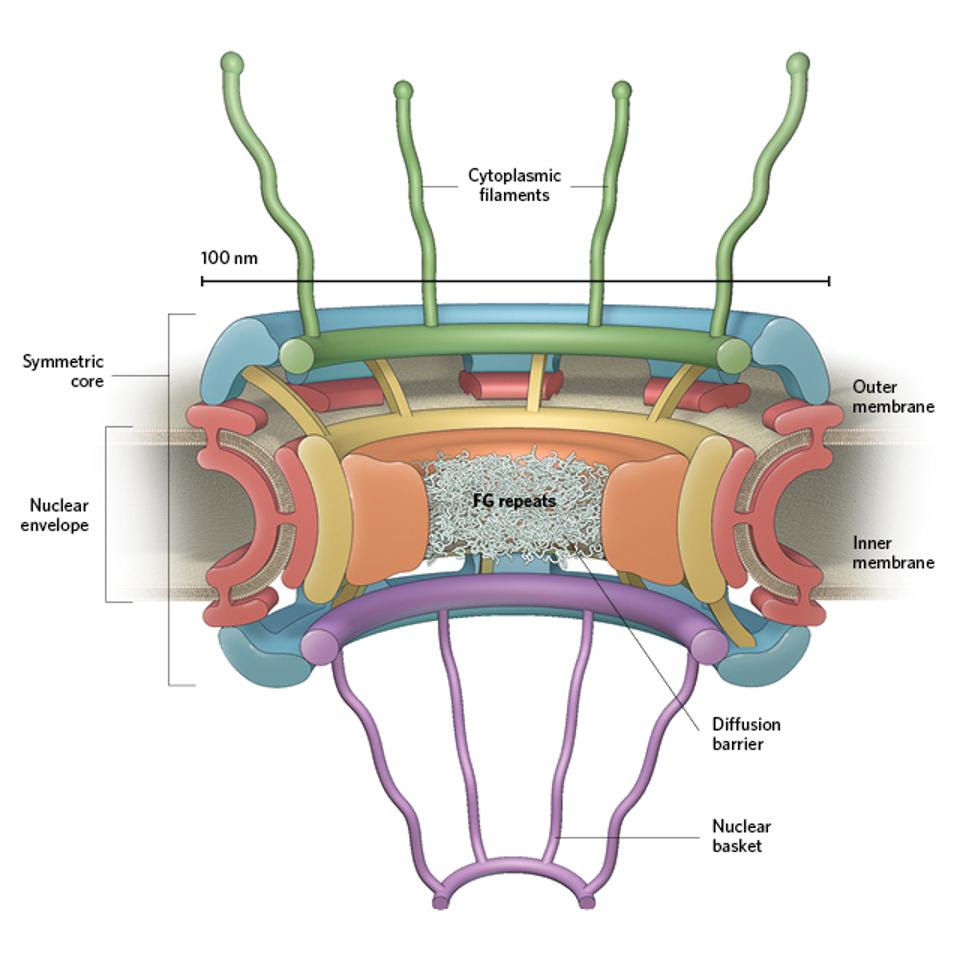
Nuclear pore complexFigure 3: Nuclear pore complexTHE SCIENTIST MAGAZINE
NSP1 blocks the export of messenger RNAs by binding to two critical components of the transport complex, nuclear RNA export factor 1 (NXF1) and NTF2-related export protein 1 (NXT1). The result is messenger RNAs, if they are made, are retained in the nucleus. If they can’t be translated, they’re marked and destroyed. Once again, nuclear export of cellular messenger RNAs and RNA-derived molecules like microRNAs is blocked.
Preferential synthesis of viral proteins: NSP1 blocking translation of cellular messenger RNAs
Last but not least, SARS-CoV-2 favors translation of its own proteins over cellular proteins. In addition to assisting with the blockage of nuclear export, NSP1 also blocks the translation of cellular RNA—while permitting the translation of viral RNA. So efficient is this gatekeeping mechanism that eight hours following infection, the majority of new proteins made are viral, not cellular. Blocking the translation of cellular messenger RNAs by NSP1 also inhibits the synthesis of interferon and any other cellular protein that might activate an immune response.
NSP1 acts by obstructing the entry of messenger RNA into the ribosome (Figure 4 & 5). A finger of NSP1 extends into the entry port of the ribosome to recognize and bind a sequence of the 18s ribosomal RNA that lies within the channel.
NSP1 blocks ribosome entry of cellular messenger RNA.Figure 4 & 5: NSP1 blocks ribosome entry of cellular messenger RNA. NSP1 permits ribosome entry of viral messenger RNA carrying a five prime loop. ACCESS HEALTH INTERNATIONAL
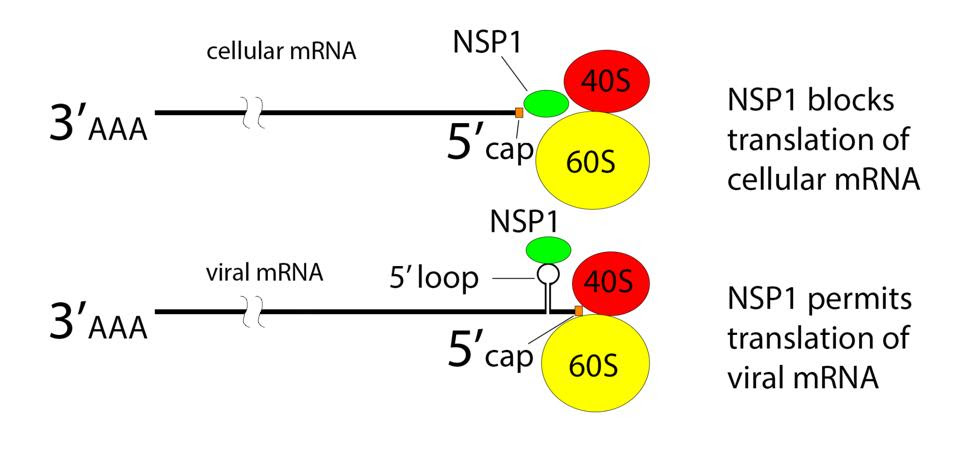
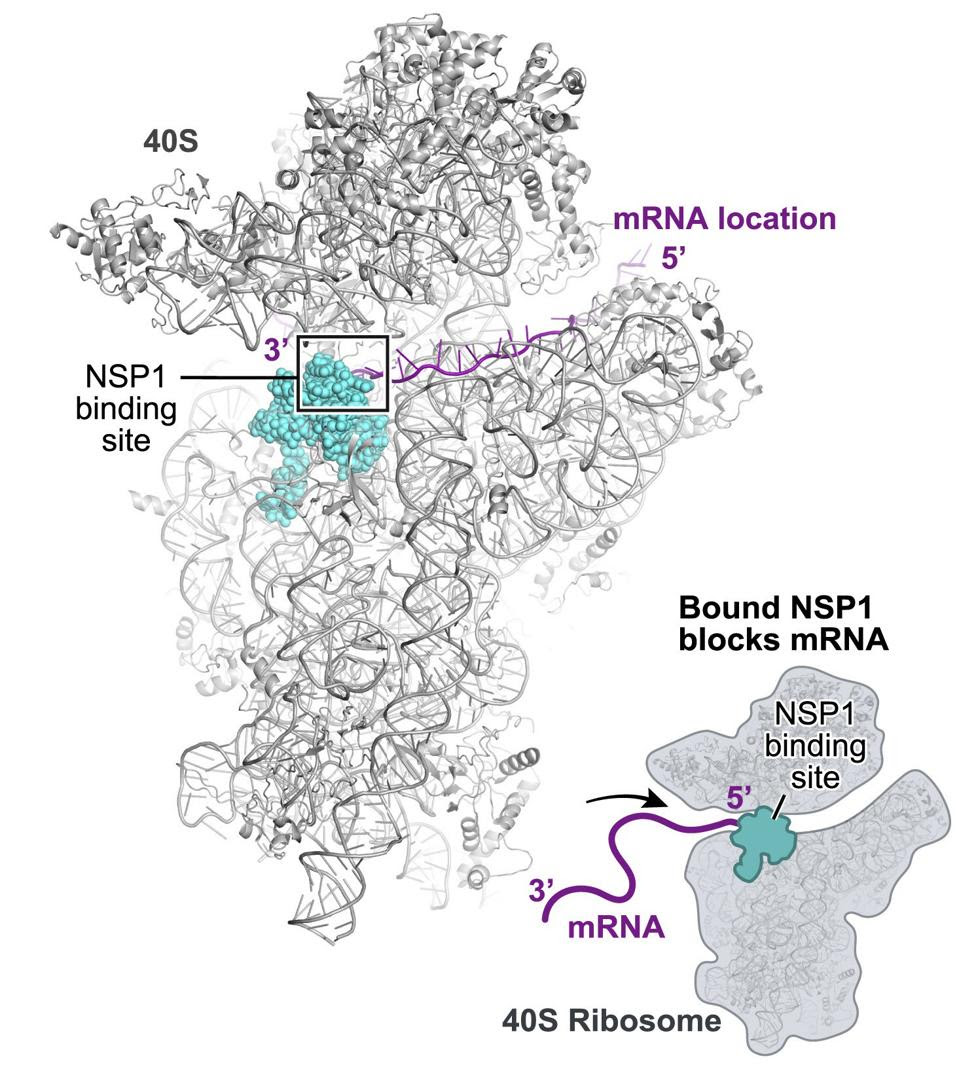
NSP1 Binds to 18S Near the mRNA Entry Channel to Suppress TranslationFigure 6: NSP1 Binds to 18S Near the mRNA Entry Channel to Suppress Translation“SARS-COV-2 DISRUPTS SPLICING, TRANSLATION, AND PROTEIN TRAFFICKING TO SUPPRESS HOST DEFENSES” HTTPS://WWW.CELL.COM/CELL/FULLTEXT/S0092-8674(20)31310-6?_RETURNURL=HTTPS%3A%2F%2FLINKINGHUB.ELSEVIER.COM%2FRETRIEVE%2FPII%2FS0092867420313106%3FSHOWALL%3DTRUE…Insert Text Above
How then are viral proteins made? A recent study offers some clues. Each viral messenger RNA shares the same 5 prime untranslated terminus. The 5 prime terminus contains several stem-loop structures. The loop structure proximal to the 5 prime end is required for translation in the presence of NSP1. Hybrid messenger RNAs that carry this 5 prime end of the genome, including the loop, are also translated in the presence of NSP1.
The location of the stem-loop structure relative to the 5 prime end is critical. The still untested hypothesis is that NSP1 recognizes the 5 prime viral messenger RNA terminus and releases blockage of the entry channel. As far as I know, this is a unique mechanism of translational regulation.
The consequence of blocking cellular messenger RNAs is not only that it blocks any newly made cellular messenger RNAs, but it blocks existing messenger RNAs as well, providing a favorable environment for viral replication. The messenger RNAs already in the cytoplasm of the cell can no longer be used. The effect of this is twofold, limiting the production of cellular proteins altogether, allowing viral RNA proteins to be made.

‘Checkpoint Charlie’ In West BerlinView of the multiple barriers at “Checkpoint Charlie” between East and West Berlin.
Summary
To recap, SARS-CoV-2 first ensures that the transcription of some cellular messenger RNAs cannot be initiated. If they are initiated, some are not spliced. If they are not spliced, they remain stuck in the nucleus. And if they exit the nucleus, some are not translated.
In part nine of this series, I will discuss how SARS-CoV-2 prevents cells from signaling their neighbors and by blocking recognition of the infected cell by immune killer cells.

