This is part of a continuing series describing antiviral antibodies to prevent and treat SARS-CoV-2 infections. In this series, we will discuss the fundamental nature of virus evolution, how SARS-CoV-2 has mutated to evade neutralizing antibodies, and our latest attempts to fight against these mutations with more recent and improved antibody candidates.
Alongside vaccines, monoclonal antibodies have been billed as the most effective means of treating and preventing Covid-19. This rang true for the first several months of the pandemic, as antibodies designed to neutralize the SARS-CoV-2 Spike protein were significantly effective. However, as the virus mutated, the potency of antibodies declined while virus immune evasion rose. To counter this trend, there is now an ongoing search for broadly neutralizing antibodies that can overcome not just a single strain but all strains of SARS-CoV-2, current and future.
In this series, we have discussed several pan-variant monoclonal antibodies, all of which show promise against current Omicron strains and previous variants of concern such as Alpha, Beta, and Delta. Here we analyze another described in a study by Luo et al.: the SP1-77 antibody.
SP1-77 Antibody Origin
The typical search for monoclonal antibodies includes the collection of sera from Covid-19 patients and isolating the often dozens or hundreds of distinct antibodies found, testing them individually on pseudoviruses with the SARS-CoV-2 Spike for binding capacity, then sorting for the best neutralizers.
A team of researchers from the Massachusetts Institute of Technology took a different approach. Rather than a blind treasure hunt in the convalescent sera of a Covid patient, the researchers developed a specialized mouse model modified with human gene segments VH1-2 and Vκ1-33. These genes are associated with complementarity-determining-region-3 (CDR3) sequences. In essence, this modification results in a far more diverse B cell response when exposed to pathogens like SARS-CoV-2, leading to the identification of more unique and distinct monoclonal antibodies to test.
Luo et al. immunized the mouse models by exposing them to the Wuhan Spike twice over four weeks. All mice involved developed strong resistance to the SARS-CoV-2 Spike, yielding 96 antibodies for the researchers to test.
SP1-77 Antibody Neuralization
Of the 96 candidates, the researchers narrowed the field down to nine that targeted the Spike protein specifically and then down to three that potently neutralized the Wuhan strain of the virus: VHH7-5-82, VHH7-7-53, and SP1-77. These three were tested for neutralization against current and previous variants of concern, both pseudotype and live viruses.
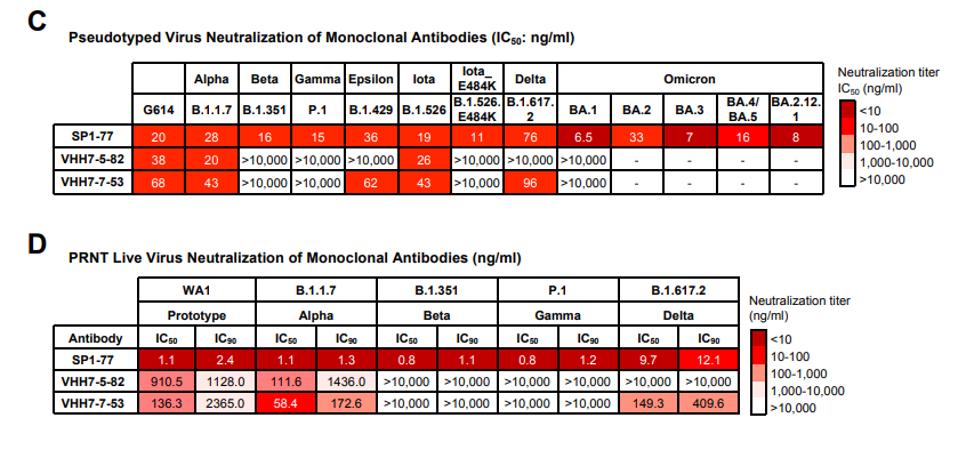
Figure one demonstrates the neutralizing prowess of SP1-77. In the pseudotyped virus assays, SP1-77 neutralized all variants of concern tested to varying degrees, including Alpha, Beta, Gamma, Delta, and both previous and current Omicron strains. Alternatively, the other two antibodies tested failed to neutralize any Omicron strains and struggled against many other previous variants of concern.
The live virus neutralization paints a similar picture. Omicron was omitted from the live virus tests for safety, but SP1-77 again neutralizes Alpha, Beta, Gamma, and Delta, whereas the others again struggle.
SP1-77 Antibody Target
A closer examination of the SP1-77 antibody via cryo-electron microscopy reveals an alternate solution to neutralization aside from inhibiting ACE2 binding. SP1-77 targets sites on the opposite side of the receptor-binding domain from the ACE2 binding site. In particular, the antibody epitope targeted positions ranging from 339-346, 440-450, and 499. In most variants of concern, these amino acids are unmutated, with the lone exception being G339 and G446 in some Omicron strains being changed.
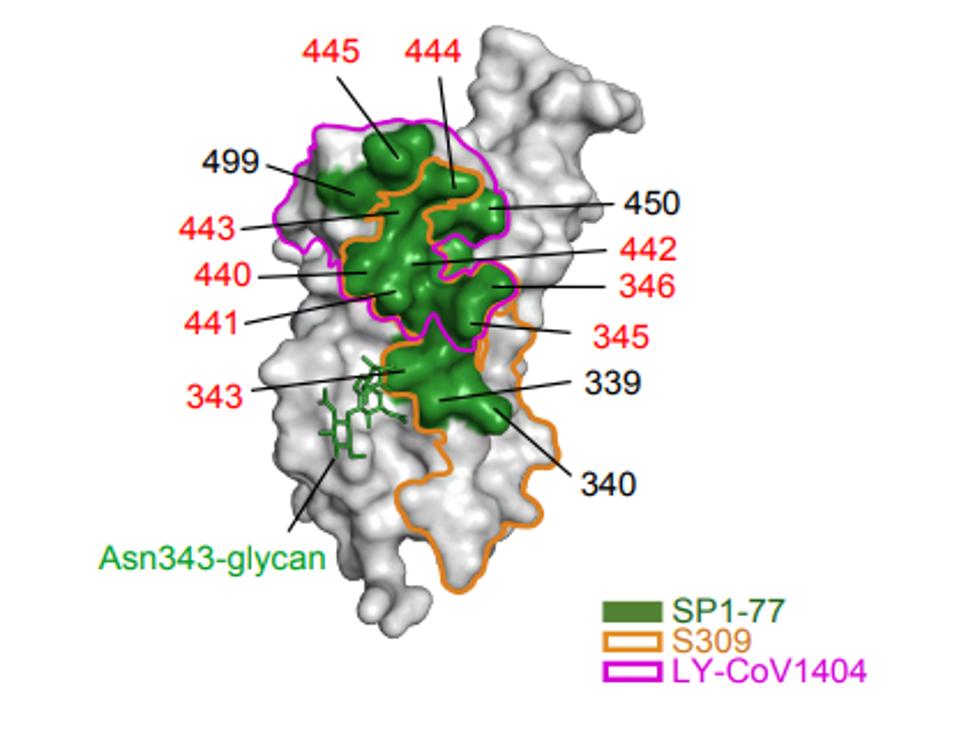
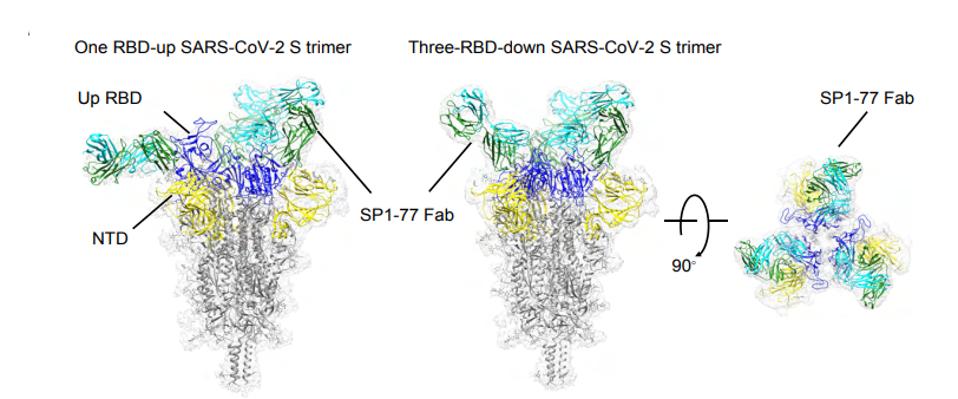
Further analysis found that the antibody binding results in either the blockage of S2’ cleavage and/or inhibition of S1 dissociation. What does this mean? After the virus binds a human ACE2 receptor, the S1 and S2 portions of the Spike must disengage for membrane fusion to occur. If the antibody is blocking either S2’ cleavage or S1 dissociation, it is inertly preventing the virus from fusing with the host cell, preventing that virus from replicating within the host cell and moving on to another.
We have previously described two other antibodies that bind to different sites. The first, 35B5, targets the N-terminal domain positions N165 and N234, which act together as a molecular switch for the Spike’s changing up and down conformations. The second, Cv2.1169, targets a different region of the receptor-binding domain than SP1-77, namely contact points including Y473, S477, T478, F486, N487, and Q493. We will describe more in the coming days and weeks.
Echoing Luo et al., SP1-77 represents an opportunity for highly potent monoclonal treatments. In addition to being an effective antibody on its own, due to its distinct residue targets, SP1-77 could be easily paired with one or two other antibodies to create a highly potent antibody cocktail for the treatment and prevention of Covid-19. Such cocktails should be immediately investigated.
Additionally, the novel humanized mouse model is a triumph of antibody discovery technology. The methods by Luo et al. should be further explored in all antibody discovery trials, potentially leading to dozens of antibodies to aid the millions continuing to get sick in this extended pandemic.